Next Generation Sequencing Workshop
Background
One of the most significant breakthroughs in genetics is the development of Next Generation Sequencing (NGS) technology. This powerful tool allows scientists to sequence millions of DNA fragments simultaneously to producing vast amounts of genomic data in a single experiment. However, NGS technology requires specialized knowledge and skills to operate effectively. Therefore, In 2019, i co-facilitated an NGS workshop to help researchers and post graduate students acquire the skills and knowledge necessary to leverage this technology in their research.
Workshop objectives
The workshop aimed to achieve the following objectives:
Help researchers identify how to utilize next generation sequencing for their own research projects
Give theoretical knowledge as well as practical experience on applying Illumina sequencing on small genomes and PCR amplicons.
This encompassed how to set up a sequencing run, prepare DNA libraries, monitor sequencing runs, troubleshoot and optimize runs and extract sequencing data and determine sample quality.
Provide guidance for researchers who were interested in utilizing the technology after the workshop concludes.
Workshop structure and activities
The NGS workshop was designed for 20 researchers who have some basic knowledge of genomics and molecular biology but have little or no experience with Illumina next generation sequencing technology.
The workshop was organized by Professor Pascal O. Bessong and Doctor Nontokozo Dephney Matume. Professor Bessong and Doctor Matume facilitated the theoretical sessions while Doctor Matume and i facilitated the practical sessions of the three day workshop hosted at Life Sciences building of University of Venda.
The workshop was structured so as to cover theoretical and practical details that went into conducting a sequencing run.
The first day of the workshop focused on the theoretical aspects of NGS technology such as the fundamental principles of NGS technology, including library preparation, sequencing, and data analysis. The talk also mentioned advantages and limitations of different NGS technology and the factors that influence the quality of sequencing data. Then two methods for setting up a sequencing run was demonstrated that is setting up a local run manager or online basespace hub. Basic calculations done during library preparation such as calculations for purifying and quantifying DNA and normalizing amplicons and DNA libraries were demonstrated.
Workshop organizers and facilitators Research Professor Pascal Bessong (person wearing brown cap) and Doctor Daphney Matume (lady standing on the podium infront of projector screen) explaining theory behind Next Generation sequencing during first day of workshop
The second day of the workshop was devoted to practical sessions. Participants were divided into groups, and each group was assigned two viral amplicons to gain hands-on experience of the steps taken during library preparation. This involved, purification of amplicons, normalizing amplicons, fragmenting and tagmenting, cleaning DNA libraries and quantifying and determining size of libraries, normalizing and pooling DNA libraries and loading pooled DNA libraries to an Illumina MiniSeq sequencing machine.
 Bixa (first person on the left of image) demonstrating to workshop attendants DNA library preparation for Next Generation sequencing
Bixa (first person on the left of image) demonstrating to workshop attendants DNA library preparation for Next Generation sequencing
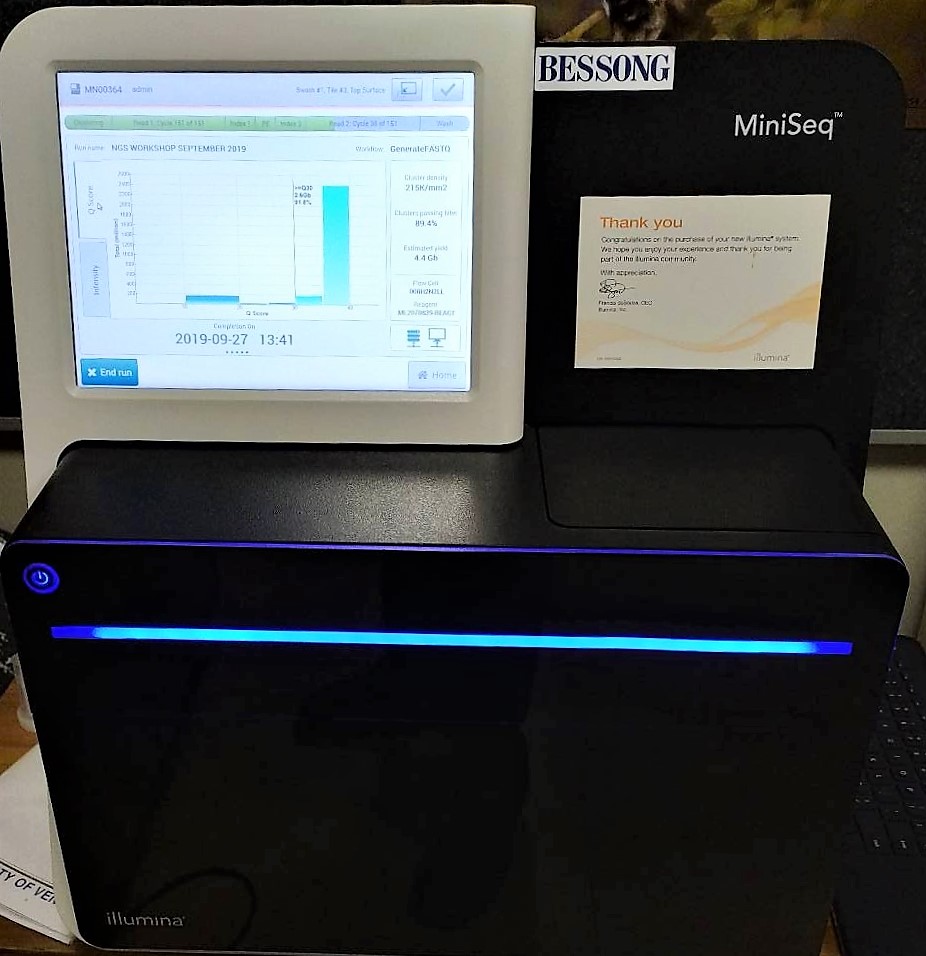 Sequencing run in progress
Sequencing run in progress
 Workshop participants posing for photo
Workshop participants posing for photo
The third day of the workshop began with demonstrating how to evaluate the run quality. Later on, Dr. Matume demonstrated how to extract sequencing data and analyzing it using Geneious. The workshop participants were provided with licences they could use on their installed Geneious software so as to get a hands on experience in performing quality control on sequenced data, map reads to a reference genome and perform variant calling.
 Doctor Matume demonstrating Next generation sequencing analysis
Doctor Matume demonstrating Next generation sequencing analysis
The workshop was highly interactive, and had robust and lively engagement from the workshop participants. The hands-on experience provided workshop participants with a practical understanding of NGS technology and its applications.
Benefits of the Workshop
The NGS workshop provided participants with a comprehensive understanding of NGS library preparation and data analysis. Participants gained hands-on experience in NGS experiments, which is essential for the successful application of NGS technology in their research. The workshop also provided an opportunity for participants to interact with other researchers and discuss their research interests and challenges.
The workshop also helped researchers understand the applications and limitations of NGS technology and how to they could leverage this technology in their research.
There was alot of interest from researchers to sequence samples from their projects and pooling samples was discussed as a cost-cutting measure.
 Group photo of workshop participants
Group photo of workshop participants

